Multiple Linear regression: Residuals
> lm2<-lm(wt~age+ht, data=d)
> summary(lm2)
Call:
lm(formula = wt ~ age + ht, data = d)
Residuals:
Min 1Q Median 3Q Max
-2.48498 -0.53548 0.01508 0.51986 2.77917
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) -8.297442 0.865929 -9.582 <2e-16 ***
age 0.005368 0.010169 0.528 0.598
ht 0.228086 0.014205 16.057 <2e-16 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error: 0.9035 on 182 degrees of freedom
Multiple R-squared: 0.9163, Adjusted R-squared: 0.9154
F-statistic: 995.8 on 2 and 182 DF, p-value: < 2.2e-16
The estimated coefficient 
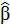 using OLS
using OLS
- What is residual denoted as ε.
Consider a multiple variate linear regression model (MvLR). Suppose that there is linear relationship between X and Y.
In another word, 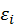
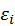 is independent random variable (iid).
is independent random variable (iid).
There are equations if there is column matrix εnx1:
So column matrix εnx1 follows multivariates Gaussian distribution: ε ~ MVG(0, σ2I), which indicates a multivariate normal distribution of ε with mean 0nx1 and the variance-covariance matrix σ2Inxn.
There is residuals sum of squares (RSS):
- Estimation of residual using OLS.
Because the population residuals are unknown, so consider OLS residuals based on observed data.
Consider OLS residuals 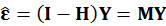

Consider another equation of 
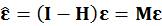
OLS residual 
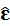 is unbiased estimator of ε.
is unbiased estimator of ε.
Proof 

Proof variance of 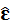
 :
: 
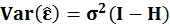
There is linear algebra theorem Var(Xv)=XVar(v)X' where v is a column and X is a matrix.
So 

- Estimation of variance of residuals
It is supposed that ε follow gaussian distribution, ε ~ MVG(0, σ2I), σ2 is the population variance, but it is unknown. We then estimate σ2 with 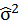
 . So there is a definition on
. So there is a definition on 
 , n is number of observations, and p is number of coefficients:
, n is number of observations, and p is number of coefficients:
Proof 
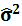 is an unbiased estimator of the variance of the disturbance.
is an unbiased estimator of the variance of the disturbance.
There is prediction error sum of squared (PESS) or residuals sum of squares (RSS):
So residuals standard error 
 :
:
95% CI of 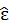
 :
:
The PESS estimate is useful as summary measure of a model's ability to predict new observations.
- R Code
R code:
#matrix of independent variables
> X<-cbind(rep(1, nrow(d)), as.matrix(d[,c('age','ht')]))
> head(X)
age ht
446 1 1 52.4
671 1 1 53.6
241 1 2 56.2
261 1 2 54.7
301 1 2 54.1
196 1 3 57.0
> dim(X)
[1] 185 3
#matrix of depencent variable:
> Y<-as.matrix(d$wt)
> dim(Y)
[1] 185 1
#degree of freedom
> (df=nrow(X)-length(coef(lm2)))
[1] 182
#OLS_beta: beta0, beta1, beta2
> (OLS_beta<-ginv(t(X)%*%X)%*%t(X)%*%Y)
[,1]
[1,] -8.297442239
[2,] 0.005368228
[3,] 0.228085501
#hat matrix
> H<-X%*%ginv(t(X)%*%X)%*%t(X)
> dim(H)
[1] 185 185
Here is R code to calculate OLS residuals:
> residuals<-(diag(1, nrow(X))-H)%*%Y
> dim(residuals)
> #standard error of residuals
> (se_residuals<-sqrt(t(Y)%*%(diag(1, nrow(X))-H)%*%Y/df))
[,1]
[1,] 0.9034802
> #95% CI of residuals
> (t95<-qt(0.975, df=182, lower.tail=F))
[1] 1.973084
> df<-data.frame('residuals'=residuals,
+ 'lower_bound'=as.numeric(residuals)-t95*se_residuals,
+ 'upper_bound'=as.numeric(residuals)+t95*se_residuals)
> head(df)
residuals lower_bound upper_bound
446 0.44039334 -1.342249 2.223036
671 -0.13330852 -1.915951 1.649334
241 0.06830038 -1.714342 1.850943
261 -0.08957137 -1.872214 1.693071
301 0.04728045 -1.735362 1.829923
196 0.48046383 -1.302179 2.263106
Then histogram of residuals
> hist(residuals, main='OLS residuals')
> abline(v=c(-t95*se_residuals, 0, t95*se_residuals),
+ col=c('blue', 'green', 'red'), lwd=3)
No comments:
Post a Comment